Inspired by biological neurons and neural circuits, we model artificial neural network architectures with biological interpretability.
 The Dendritic Neural Model (DNM) is a novel neural model with plastic dendritic morphology that we have proposed.
The Dendritic Neural Model (DNM) is a novel neural model with plastic dendritic morphology that we have proposed.
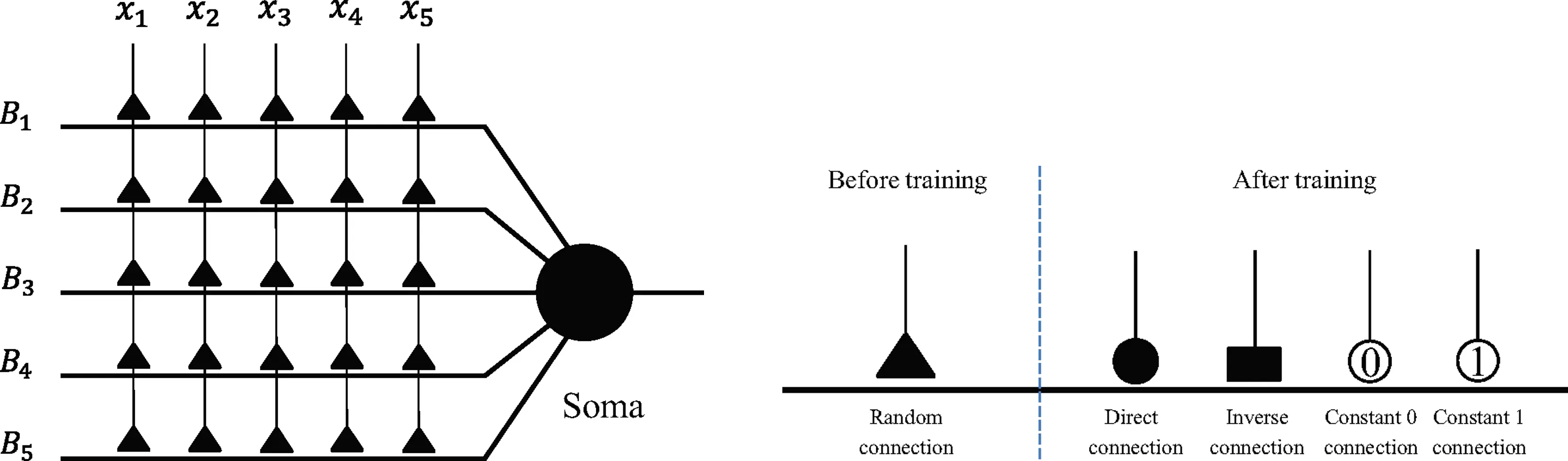 The DNM consists of synaptic layer, dendritic layer, membrane layer, and soma, with each layer performing corresponding neural functions through different activation functions.
The DNM consists of synaptic layer, dendritic layer, membrane layer, and soma, with each layer performing corresponding neural functions through different activation functions.
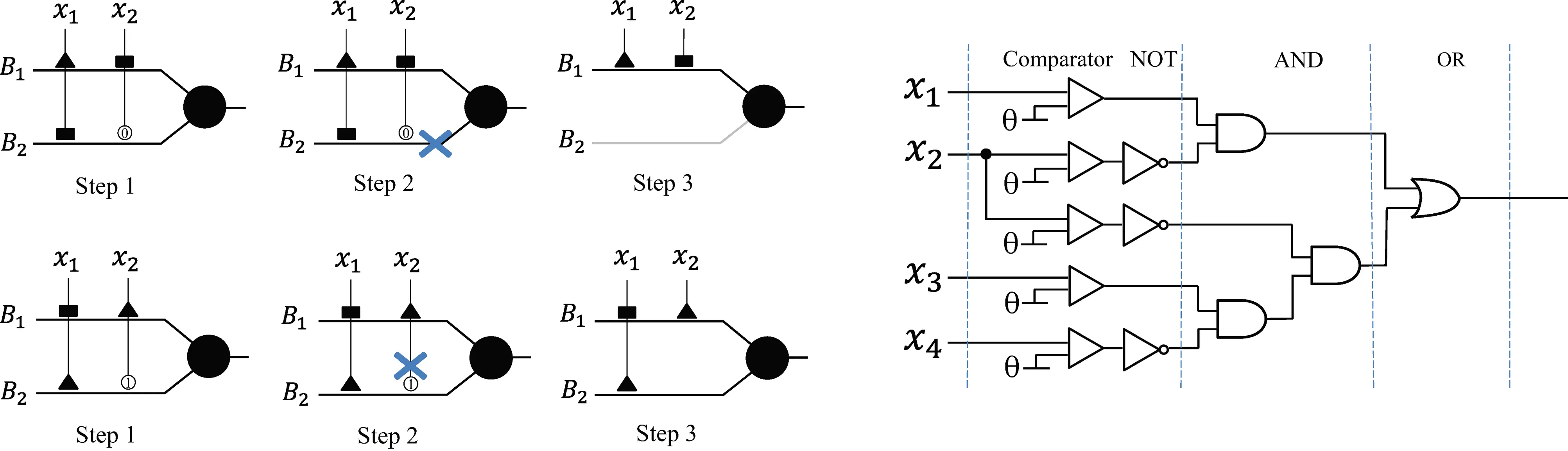 Through a neural pruning scheme, DNM can eliminate redundant synapses and dendritic branches to simplify its architecture and form unique neuronal morphologies for each specific task.
Through a neural pruning scheme, DNM can eliminate redundant synapses and dendritic branches to simplify its architecture and form unique neuronal morphologies for each specific task.
Through a logic approximation scheme, DNM can be converted into a Logic Circuit Classifier (LCC), which consists only of comparators and logic AND, OR, and NOT gates.
LCC is easy to implement for large-scale parallel computing on hardware, such as Field-Programmable Gate Arrays (FPGA) and Very Large Scale Integration (VLSI).
Yajiao Tang, Zhenyu Song, Yulin Zhu, Maozhang Hou, Cheng Tang, and Junkai Ji. “Adopting a dendritic neural model for predicting stock price index movement.” Expert Systems with Applications (2022): 117637. View Paper
Cheng Tang, Junkai Ji, Qiuzhen Lin, and Yan Zhou. “Evolutionary Neural Architecture Design of Liquid State Machine for Image Classification.” ICASSP 2022-2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2022. View Paper
Junkai Ji, Jiajun Zhao, Qiuzhen Lin, and Kay Chen Tan. “Competitive Decomposition-Based Multiobjective Architecture Search for the Dendritic Neural Model.” IEEE Transactions on Cybernetics (2022). View Paper
Junkai Ji, Cheng Tang, Jiajun Zhao, Zheng Tang, and Yuki Todo. “A Survey on Dendritic Neuron Model: Mechanisms, Algorithms and Practical Applications.” Neurocomputing (2022). View Paper
Junkai Ji, Minhui Dong, Qiuzhen Lin, and Kay Chen Tan. “Noninvasive Cuffless Blood Pressure Estimation With Dendritic Neural Regression.” IEEE Transactions on Cybernetics (2022). View Paper
Cheng Tang, Yuki Todo, Junkai Ji, and Zheng Tang. “A novel motion direction detection mechanism based on dendritic computation of direction-selective ganglion cells.” Knowledge-Based Systems (2022): 108205. View Paper
Zhenyu Song, Cheng Tang, Junkai Ji, Yuki Todo, and Zheng Tang. “A Simple Dendritic Neural Network Model-Based Approach for Daily PM2. 5 Concentration Prediction.” Electronics 10, no. 4 (2021): 373. View Paper
Minhui Dong, Cheng Tang, Junkai Ji, Qiuzhen Lin, and Ka-Chun Wong. “Transmission trend of the COVID-19 pandemic predicted by dendritic neural regression.” Applied Soft Computing 111 (2021): 107683. View Paper
Shangce Gao, Mengchu Zhou, Ziqian Wang, Daiki Sugiyama, Jiujun Cheng, Jiahai Wang, and Yuki Todo. “Fully complex-valued dendritic neuron model.” IEEE Transactions on Neural Networks and Learning Systems (2021). View Paper
Xiliang Zhang, Yuki Todo, Cheng Tang, and Zheng Tang. “The Mechanism of Orientation Detection Based on Dendritic Neuron.” 2021 IEEE 4th International Conference on Big Data and Artificial Intelligence (BDAI). IEEE, 2021. View Paper
Jiajun Zhao, Qiuzhen Lin, and Junkai Ji. “Network Intrusion Detection by an Approximate Logic Neural Model.” 2021 IEEE International Symposium on Software Reliability Engineering Workshops (ISSREW). IEEE, 2021. View Paper
Cheng Tang, Zhenyu Song, Yajiao Tang, Huimei Tang, Yuxi Wang, and Junkai Ji. “An Evolutionary Neuron Model with Dendritic Computation for Classification and Prediction.” International Conference on Intelligent Computing. Springer, Cham, 2021. View Paper
Junkai Ji, Minhui Dong, Qiuzhen Lin, and Kay Chen Tan. “Forecasting Wind Speed Time Series Via Dendritic Neural Regression.” IEEE Computational Intelligence Magazine 16.3 (2021): 50-66. View Paper
Zhenyu Song, Cheng Tang, Jin Qian, Bin Zhang, and Yuki Todo. “Air Quality Estimation Using Dendritic Neural Regression with Scale-Free Network-Based Differential Evolution.” Atmosphere 12.12 (2021): 1647. View Paper
Shuangbao Song, Xingqian Chen, Shuangyu Song, and Yuki Todo. “A neuron model with dendrite morphology for classification.” Electronics 10.9 (2021): 1062. View Paper
Junkai Ji, Yajiao Tang, Lijia Ma, Jianqiang Li, Qiuzhen Lin, Zheng Tang, and Yuki Todo. “Accuracy Versus Simplification in an Approximate Logic Neural Model.” IEEE Transactions on Neural Networks and Learning Systems (2020). View Paper
Cheng Tang, Junkai Ji, Yajiao Tang, Shangce Gao, Zheng Tang, and Yuki Todo. “A novel machine learning technique for computer-aided diagnosis.” Engineering Applications of Artificial Intelligence 92 (2020): 103627. View Paper
Zhenyu Song, Yajiao Tang, Junkai Ji, and Yuki Todo. “Evaluating a dendritic neuron model for wind speed forecasting.” Knowledge-Based Systems 201 (2020): 106052. View Paper
Xiaoxiao Qian, Cheng Tang, Yuki Todo, Qiuzhen Lin, and Junkai Ji. “Evolutionary Dendritic Neural Model for Classification Problems.” Complexity 2020 (2020). View Paper
Jiajun Zhao, Minhui Dong, Cheng Tang, Junkai Ji, and Ying He. “Improving Approximate Logic Neuron Model by Means of a Novel Learning Algorithm.” In International Conference on Intelligent Computing, pp. 484-496. Springer, Cham, 2020. View Paper
Junkai Ji, Minhui Dong, Cheng Tang, Jiajun Zhao, and Shuangbao Song. “A Novel Plastic Neural Model with Dendritic Computation for Classification Problems.” In International Conference on Intelligent Computing, pp. 471-483. Springer, Cham, 2020. View Paper
Zhenyu Song, Tianle Zhou, Xuemei Yan, Cheng Tang, and Junkai Ji. “Wind Speed Time Series Prediction Using a Single Dendritic Neuron Model.” In 2020 2nd International Conference on Machine Learning, Big Data and Business Intelligence (MLBDBI), pp. 140-144. IEEE, 2020. View Paper
Junkai Ji, Shuangbao Song, Yajiao Tang, Shangce Gao, Zheng Tang, and Yuki Todo. “Approximate logic neuron model trained by states of matter search algorithm.” Knowledge-Based Systems 163 (2019): 120-130. View Paper
Yuki Todo, Zheng Tang, Hiroyoshi Todo, Junkai Ji, and Kazuya Yamashita. “Neurons with multiplicative interactions of nonlinear synapses.” International journal of neural systems 29, no. 08 (2019): 1950012. View Paper
Tang, Yajiao, Junkai Ji, Yulin Zhu, Shangce Gao, Zheng Tang, and Yuki Todo. “A differential evolution-oriented pruning neural network model for bankruptcy prediction.” Complexity 2019 (2019). View Paper
Shuangyu Song, Xingqian Chen, Cheng Tang, Shuangbao Song, Zheng Tang, and Yuki Todo. “Training an approximate logic dendritic neuron model using social learning particle swarm optimization algorithm.” IEEE Access 7 (2019): 141947-141959. View Paper
Jingyao Wu, Jiaxin He, and Yuki Todo. “The dendritic neuron model is a universal approximator.” In 2019 6th International Conference on Systems and Informatics (ICSAI), pp. 589-594. IEEE, 2019. View Paper
Fei Teng, and Yuki Todo. “Dendritic Neuron Model and Its Capability of Approximation.” In 2019 6th International Conference on Systems and Informatics (ICSAI), pp. 542-546. IEEE, 2019. View Paper
Jiaxin He, Jingyao Wu, Guangchi Yuan, and Yuki Todo. “Dendritic Branches of DNM Help to Improve Approximation accuracy.” In 2019 6th International Conference on Systems and Informatics (ICSAI), pp. 533-541. IEEE, 2019. View Paper
Junkai Ji. “The improvement and hybridization of artificial neural networks and swarm intelligence.” Doctoral dissertation, University of Toyama, 2018. View Paper
Tao Jiang, Shangce Gao, Dizhou Wang, Junkai Ji, Yuki Todo, and Zheng Tang. “A neuron model with synaptic nonlinearities in a dendritic tree for liver disorders.” IEEJ Transactions on Electrical and Electronic Engineering 12, no. 1 (2017): 105-115. View Paper
Wei Chen, Jian Sun, Shangce Gao, Jiu-Jun Cheng, Jiahai Wang, and Yuki Todo. “Using a single dendritic neuron to forecast tourist arrivals to Japan.” IEICE Transactions on Information and Systems 100, no. 1 (2017): 190-202. View Paper
Ying Yu, Yirui Wang, Shangce Gao, and Zheng Tang. “Statistical modeling and prediction for tourism economy using dendritic neural network.” Computational intelligence and neuroscience 2017 (2017). View Paper
Junkai Ji, Shangce Gao, Jiujun Cheng, Zheng Tang, and Yuki Todo. “An approximate logic neuron model with a dendritic structure,” Neurocomputing 173 (2016): 1775-1783. View Paper
Tianle Zhou, Shangce Gao, Jiahai Wang, Chaoyi Chu, Yuki Todo, and Zheng Tang. “Financial time series prediction using a dendritic neuron model.” Knowledge-Based Systems 105 (2016): 214-224. View Paper
Junkai Ji, Zhenyu Song, Yajiao Tang, Tao Jiang, and Shangce Gao. “Training a dendritic neural model with genetic algorithm for classification problems.” In 2016 International Conference on Progress in Informatics and Computing (PIC), pp. 47-50. IEEE, 2016. View Paper
Zijun Sha, Lin Hu, Yuki Todo, Junkai Ji, Shangce Gao, and Zheng Tang. “A breast cancer classifier using a neuron model with dendritic nonlinearity.” IEICE Transactions on Information and Systems 98, no. 7 (2015): 1365-1376. View Paper
Tao Jiang, Dizhou Wang, Junkai Ji, Yuki Todo, and Shangce Gao. “Single dendritic neuron with nonlinear computation capacity: A case study on xor problem.” In 2015 IEEE International Conference on Progress in Informatics and Computing (PIC), pp. 20-24. IEEE, 2015. View Paper
Yuki Todo, Hiroki Tamura, Kazuya Yamashita, and Zheng Tang. “Unsupervised learnable neuron model with nonlinear interaction on dendrites.” Neural Networks 60 (2014): 96-103. View Paper
Zheng Tang, Hiroki Tamura, Makoto Kuratu, Okihiko Ishizuka, and Koichi Tanno. “A model of the neuron based on dendrite mechanisms.” Electronics and Communications in Japan (Part III: Fundamental Electronic Science) 84, no. 8 (2001): 11-24. View Paper
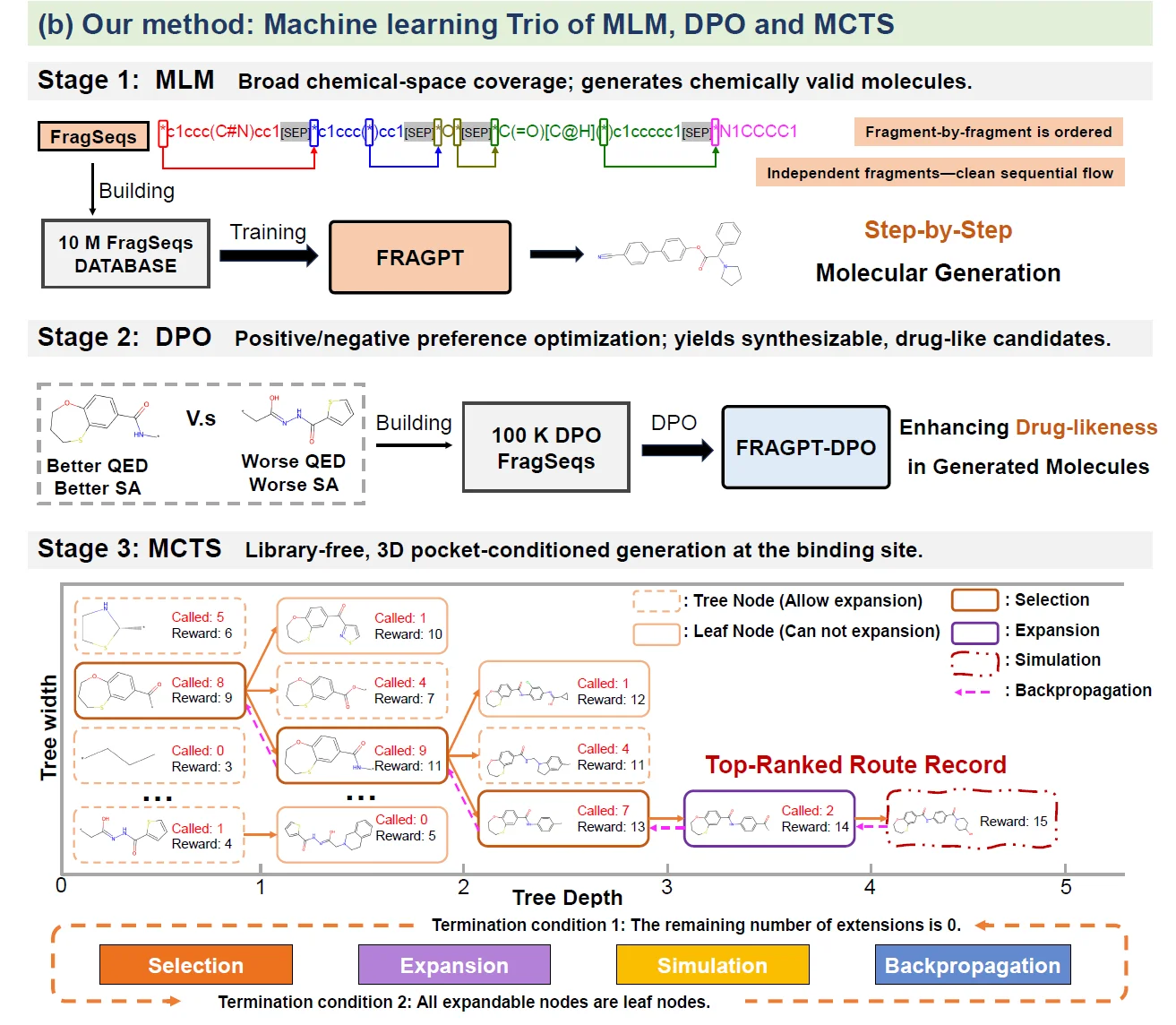
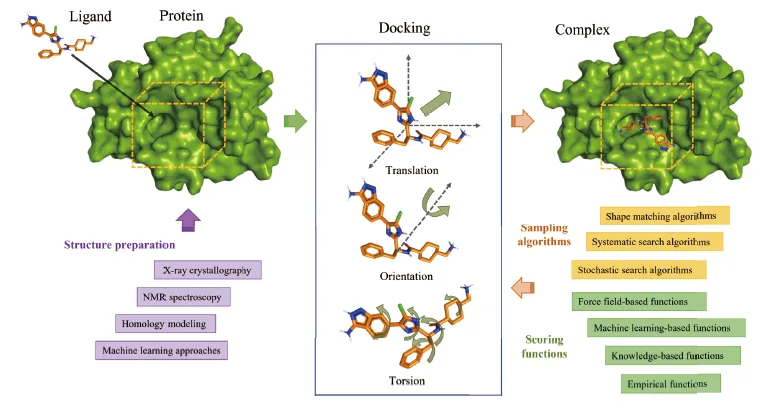
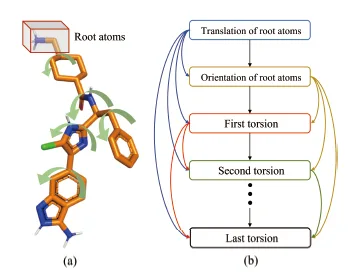
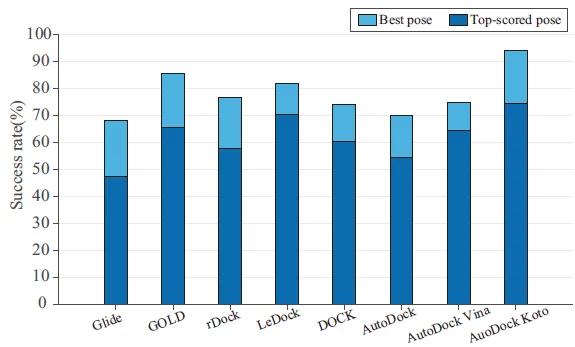
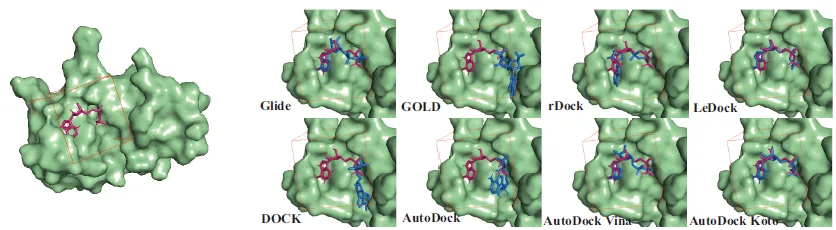
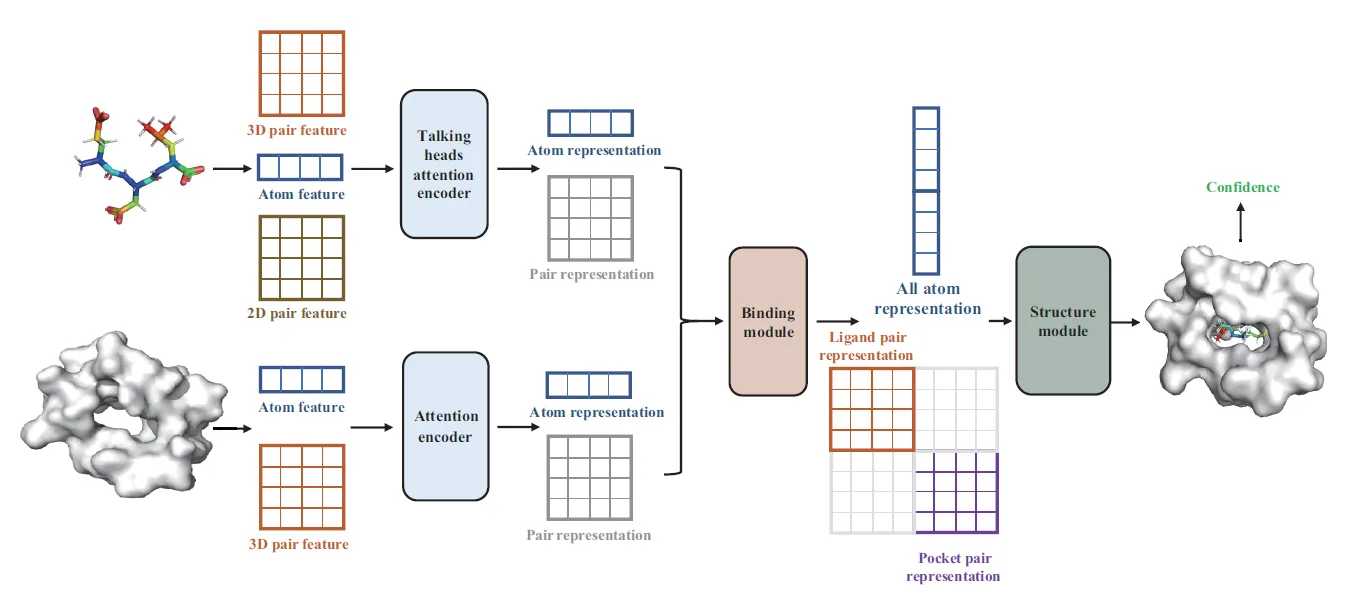
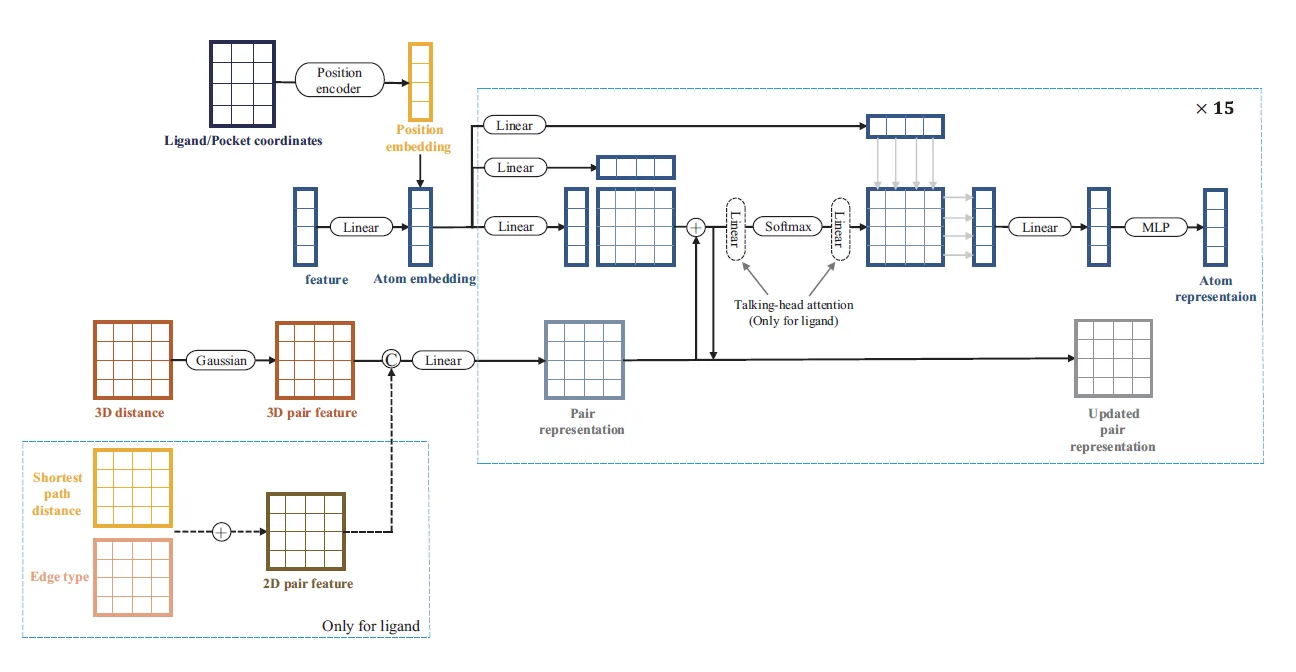
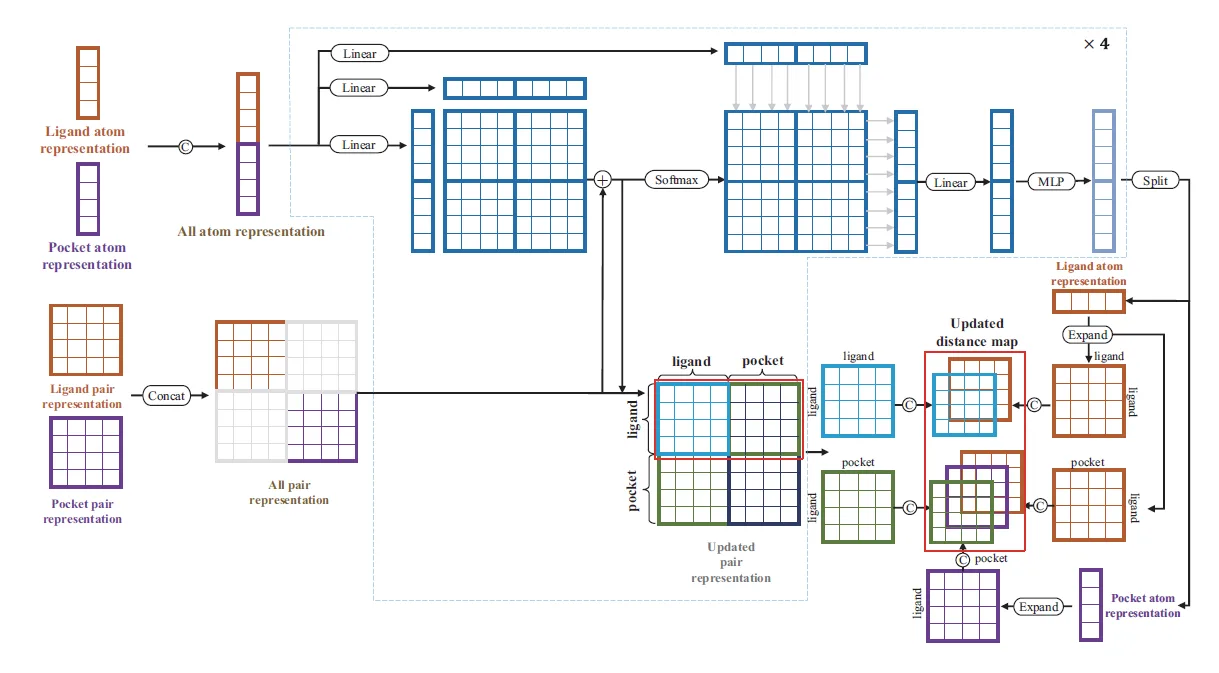
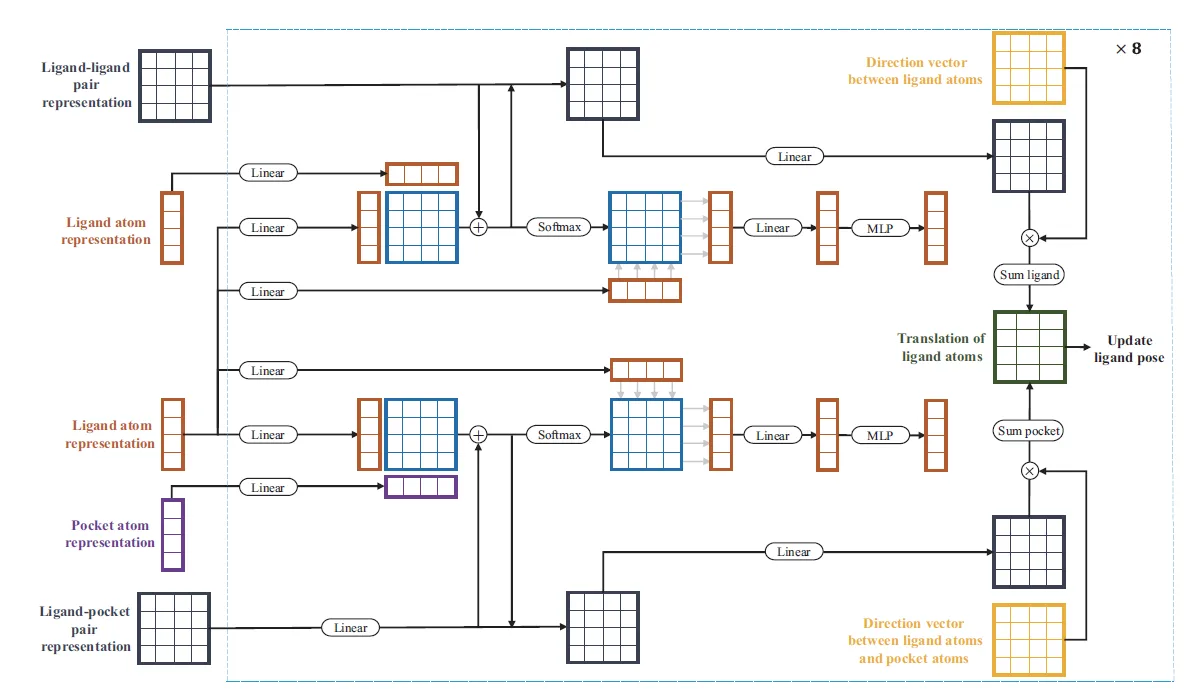
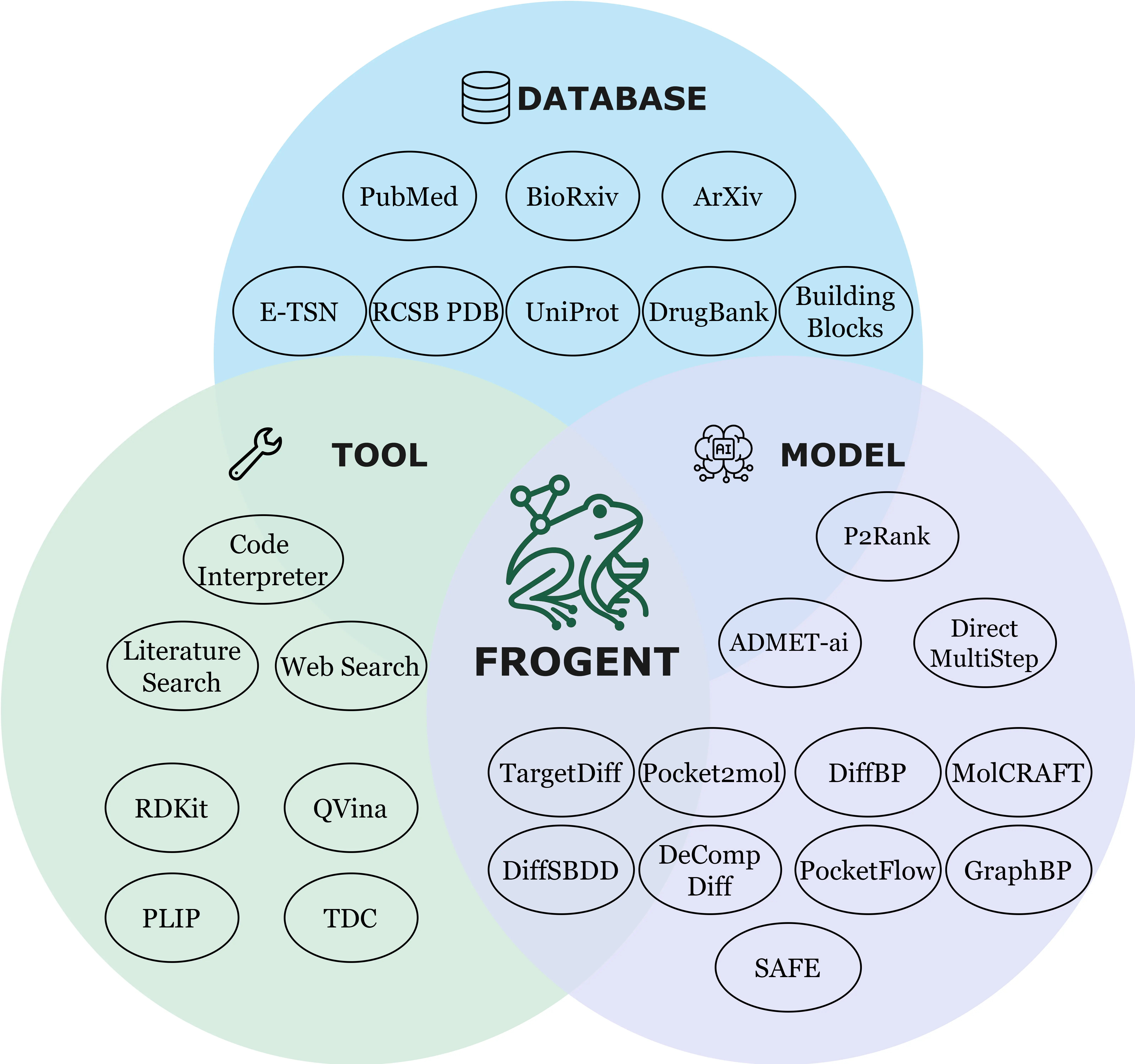
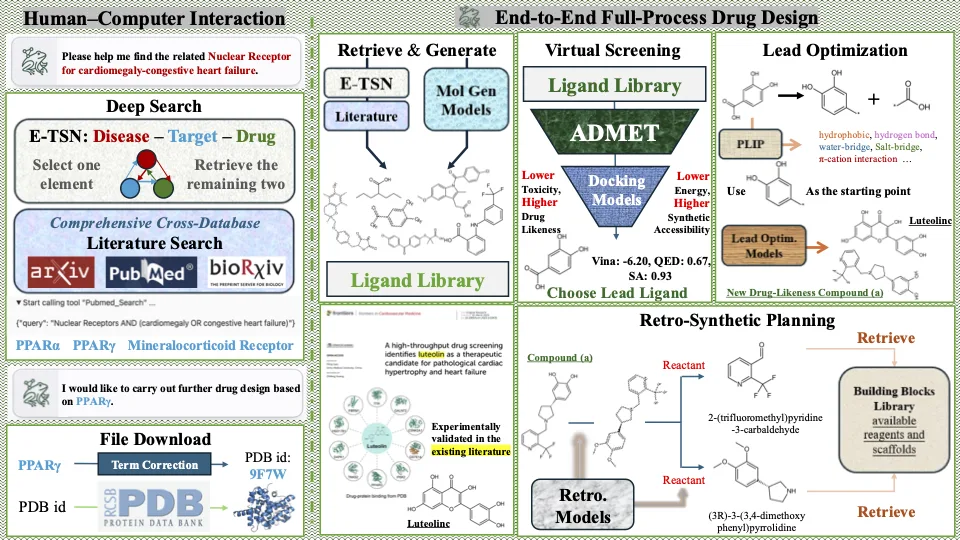
 The Dendritic Neural Model (DNM) is a novel neural model with plastic dendritic morphology that we have proposed.
The Dendritic Neural Model (DNM) is a novel neural model with plastic dendritic morphology that we have proposed.
 The DNM consists of synaptic layer, dendritic layer, membrane layer, and soma, with each layer performing corresponding neural functions through different activation functions.
The DNM consists of synaptic layer, dendritic layer, membrane layer, and soma, with each layer performing corresponding neural functions through different activation functions.
 Through a neural pruning scheme, DNM can eliminate redundant synapses and dendritic branches to simplify its architecture and form unique neuronal morphologies for each specific task.
Through a neural pruning scheme, DNM can eliminate redundant synapses and dendritic branches to simplify its architecture and form unique neuronal morphologies for each specific task.